Jichen Yang
Diagnostic Impact of Cine Clips for Thyroid Nodule Assessment on Ultrasound
Feb 01, 2026Abstract:Background: Thyroid ultrasound is commonly performed using a combination of static images and cine clips (video recordings). However, the exact utility and impact of cine images remains unknown. This study aimed to evaluate the impact of cine imaging on accuracy and consistency of thyroid nodule assessment, using the American College of Radiology Thyroid Reporting and Data System (ACR TI-RADS). Methods: 50 benign and 50 malignant thyroid nodules with cytopathology results were included. A reader study with 4 specialty-trained radiologists was then conducted over 3 rounds, assessing only static images in the first two rounds and both static and cine images in the third round. TI-RADS scores and the consequent management recommendations were then evaluated by comparing them to the malignancy status of the nodules. Results: Mean sensitivity for malignancy detection was 0.65 for static images and 0.67 with both static and cine images (p>0.5). Specificity was 0.20 for static images and 0.22 with both static and cine images (p>0.5). Management recommendations were similar with and without cine images. Intrareader agreement on feature assignments remained consistent across all rounds, though TI-RADS point totals were slightly higher with cine images. Conclusion: The inclusion of cine imaging for thyroid nodule assessment on ultrasound did not significantly change diagnostic performance. Current practice guidelines, which do not mandate cine imaging, are sufficient for accurate diagnosis.
Effectiveness of Automatically Curated Dataset in Thyroid Nodules Classification Algorithms Using Deep Learning
Feb 01, 2026Abstract:The diagnosis of thyroid nodule cancers commonly utilizes ultrasound images. Several studies showed that deep learning algorithms designed to classify benign and malignant thyroid nodules could match radiologists' performance. However, data availability for training deep learning models is often limited due to the significant effort required to curate such datasets. The previous study proposed a method to curate thyroid nodule datasets automatically. It was tested to have a 63% yield rate and 83% accuracy. However, the usefulness of the generated data for training deep learning models remains unknown. In this study, we conducted experiments to determine whether using a automatically-curated dataset improves deep learning algorithms' performance. We trained deep learning models on the manually annotated and automatically-curated datasets. We also trained with a smaller subset of the automatically-curated dataset that has higher accuracy to explore the optimum usage of such dataset. As a result, the deep learning model trained on the manually selected dataset has an AUC of 0.643 (95% confidence interval [CI]: 0.62, 0.66). It is significantly lower than the AUC of the 6automatically-curated dataset trained deep learning model, 0.694 (95% confidence interval [CI]: 0.67, 0.73, P < .001). The AUC of the accurate subset trained deep learning model is 0.689 (95% confidence interval [CI]: 0.66, 0.72, P > .43), which is insignificantly worse than the AUC of the full automatically-curated dataset. In conclusion, we showed that using a automatically-curated dataset can substantially increase the performance of deep learning algorithms, and it is suggested to use all the data rather than only using the accurate subset.
BreastSegNet: Multi-label Segmentation of Breast MRI
Jul 18, 2025Abstract:Breast MRI provides high-resolution imaging critical for breast cancer screening and preoperative staging. However, existing segmentation methods for breast MRI remain limited in scope, often focusing on only a few anatomical structures, such as fibroglandular tissue or tumors, and do not cover the full range of tissues seen in scans. This narrows their utility for quantitative analysis. In this study, we present BreastSegNet, a multi-label segmentation algorithm for breast MRI that covers nine anatomical labels: fibroglandular tissue (FGT), vessel, muscle, bone, lesion, lymph node, heart, liver, and implant. We manually annotated a large set of 1123 MRI slices capturing these structures with detailed review and correction from an expert radiologist. Additionally, we benchmark nine segmentation models, including U-Net, SwinUNet, UNet++, SAM, MedSAM, and nnU-Net with multiple ResNet-based encoders. Among them, nnU-Net ResEncM achieves the highest average Dice scores of 0.694 across all labels. It performs especially well on heart, liver, muscle, FGT, and bone, with Dice scores exceeding 0.73, and approaching 0.90 for heart and liver. All model code and weights are publicly available, and we plan to release the data at a later date.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
Segment anything model 2: an application to 2D and 3D medical images
Aug 06, 2024Abstract:Segment Anything Model (SAM) has gained significant attention because of its ability to segment varous objects in images given a prompt. The recently developed SAM 2 has extended this ability to video inputs. This opens an opportunity to apply SAM to 3D images, one of the fundamental tasks in the medical imaging field. In this paper, we extensively evaluate SAM 2's ability to segment both 2D and 3D medical images by first collecting 18 medical imaging datasets, including common 3D modalities such as computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) as well as 2D modalities such as X-ray and ultrasound. Two evaluation pipelines of SAM 2 are considered: (1) multi-frame 3D segmentation, where prompts are provided to one or multiple slice(s) selected from the volume, and (2) single-frame 2D segmentation, where prompts are provided to each slice. The former is only applicable to 3D modalities, while the latter applies to both 2D and 3D modalities. Our results show that SAM 2 exhibits similar performance as SAM under single-frame 2D segmentation, and has variable performance under multi-frame 3D segmentation depending on the choices of slices to annotate, the direction of the propagation, the predictions utilized during the propagation, etc.
How to build the best medical image segmentation algorithm using foundation models: a comprehensive empirical study with Segment Anything Model
Apr 15, 2024Abstract:Automated segmentation is a fundamental medical image analysis task, which enjoys significant advances due to the advent of deep learning. While foundation models have been useful in natural language processing and some vision tasks for some time, the foundation model developed with image segmentation in mind - Segment Anything Model (SAM) - has been developed only recently and has shown similar promise. However, there are still no systematic analyses or ``best-practice'' guidelines for optimal fine-tuning of SAM for medical image segmentation. This work summarizes existing fine-tuning strategies with various backbone architectures, model components, and fine-tuning algorithms across 18 combinations, and evaluates them on 17 datasets covering all common radiology modalities. Our study reveals that (1) fine-tuning SAM leads to slightly better performance than previous segmentation methods, (2) fine-tuning strategies that use parameter-efficient learning in both the encoder and decoder are superior to other strategies, (3) network architecture has a small impact on final performance, (4) further training SAM with self-supervised learning can improve final model performance. We also demonstrate the ineffectiveness of some methods popular in the literature and further expand our experiments into few-shot and prompt-based settings. Lastly, we released our code and MRI-specific fine-tuned weights, which consistently obtained superior performance over the original SAM, at https://github.com/mazurowski-lab/finetune-SAM.
SegmentAnyBone: A Universal Model that Segments Any Bone at Any Location on MRI
Jan 23, 2024



Abstract:Magnetic Resonance Imaging (MRI) is pivotal in radiology, offering non-invasive and high-quality insights into the human body. Precise segmentation of MRIs into different organs and tissues would be highly beneficial since it would allow for a higher level of understanding of the image content and enable important measurements, which are essential for accurate diagnosis and effective treatment planning. Specifically, segmenting bones in MRI would allow for more quantitative assessments of musculoskeletal conditions, while such assessments are largely absent in current radiological practice. The difficulty of bone MRI segmentation is illustrated by the fact that limited algorithms are publicly available for use, and those contained in the literature typically address a specific anatomic area. In our study, we propose a versatile, publicly available deep-learning model for bone segmentation in MRI across multiple standard MRI locations. The proposed model can operate in two modes: fully automated segmentation and prompt-based segmentation. Our contributions include (1) collecting and annotating a new MRI dataset across various MRI protocols, encompassing over 300 annotated volumes and 8485 annotated slices across diverse anatomic regions; (2) investigating several standard network architectures and strategies for automated segmentation; (3) introducing SegmentAnyBone, an innovative foundational model-based approach that extends Segment Anything Model (SAM); (4) comparative analysis of our algorithm and previous approaches; and (5) generalization analysis of our algorithm across different anatomical locations and MRI sequences, as well as an external dataset. We publicly release our model at https://github.com/mazurowski-lab/SegmentAnyBone.
Adaptive-avg-pooling based Attention Vision Transformer for Face Anti-spoofing
Jan 10, 2024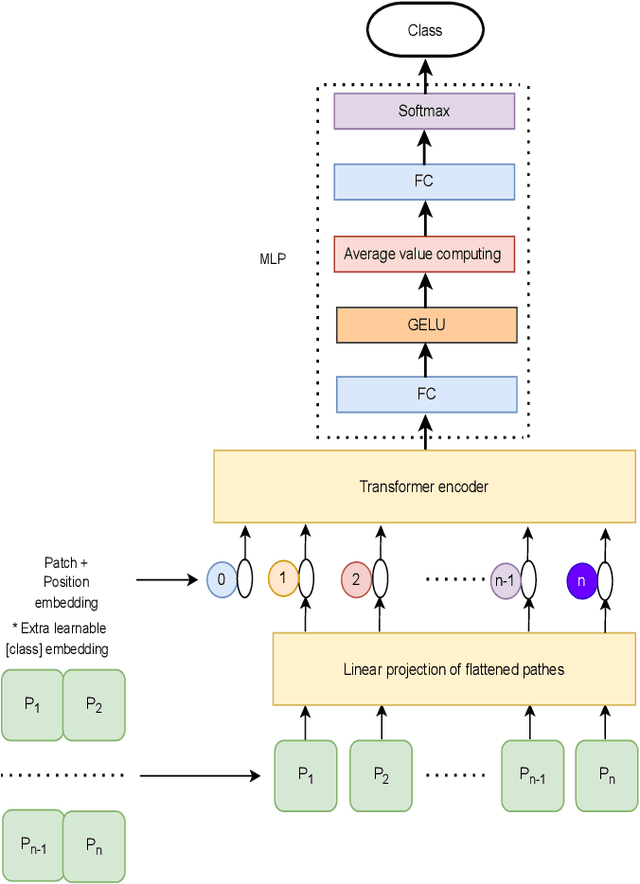
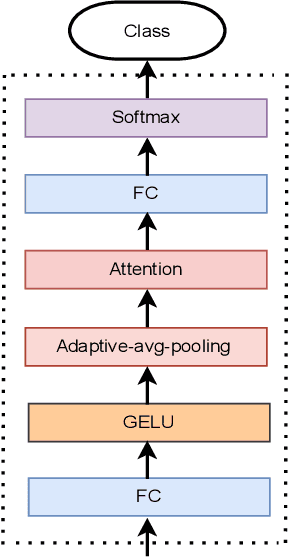
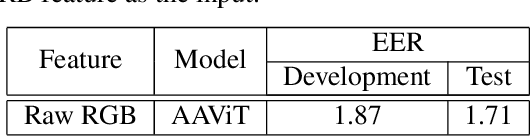
Abstract:Traditional vision transformer consists of two parts: transformer encoder and multi-layer perception (MLP). The former plays the role of feature learning to obtain better representation, while the latter plays the role of classification. Here, the MLP is constituted of two fully connected (FC) layers, average value computing, FC layer and softmax layer. However, due to the use of average value computing module, some useful information may get lost, which we plan to preserve by the use of alternative framework. In this work, we propose a novel vision transformer referred to as adaptive-avg-pooling based attention vision transformer (AAViT) that uses modules of adaptive average pooling and attention to replace the module of average value computing. We explore the proposed AAViT for the studies on face anti-spoofing using Replay-Attack database. The experiments show that the AAViT outperforms vision transformer in face anti-spoofing by producing a reduced equal error rate. In addition, we found that the proposed AAViT can perform much better than some commonly used neural networks such as ResNet and some other known systems on the Replay-Attack corpus.
Segment Anything Model for Medical Image Analysis: an Experimental Study
Apr 25, 2023



Abstract:Training segmentation models for medical images continues to be challenging due to the limited availability and acquisition expense of data annotations. Segment Anything Model (SAM) is a foundation model trained on over 1 billion annotations, predominantly for natural images, that is intended to be able to segment the user-defined object of interest in an interactive manner. Despite its impressive performance on natural images, it is unclear how the model is affected when shifting to medical image domains. Here, we perform an extensive evaluation of SAM's ability to segment medical images on a collection of 11 medical imaging datasets from various modalities and anatomies. In our experiments, we generated point prompts using a standard method that simulates interactive segmentation. Experimental results show that SAM's performance based on single prompts highly varies depending on the task and the dataset, i.e., from 0.1135 for a spine MRI dataset to 0.8650 for a hip x-ray dataset, evaluated by IoU. Performance appears to be high for tasks including well-circumscribed objects with unambiguous prompts and poorer in many other scenarios such as segmentation of tumors. When multiple prompts are provided, performance improves only slightly overall, but more so for datasets where the object is not contiguous. An additional comparison to RITM showed a much better performance of SAM for one prompt but a similar performance of the two methods for a larger number of prompts. We conclude that SAM shows impressive performance for some datasets given the zero-shot learning setup but poor to moderate performance for multiple other datasets. While SAM as a model and as a learning paradigm might be impactful in the medical imaging domain, extensive research is needed to identify the proper ways of adapting it in this domain.
Deep Learning for Classification of Thyroid Nodules on Ultrasound: Validation on an Independent Dataset
Jul 27, 2022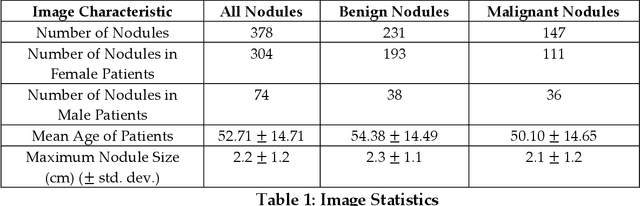
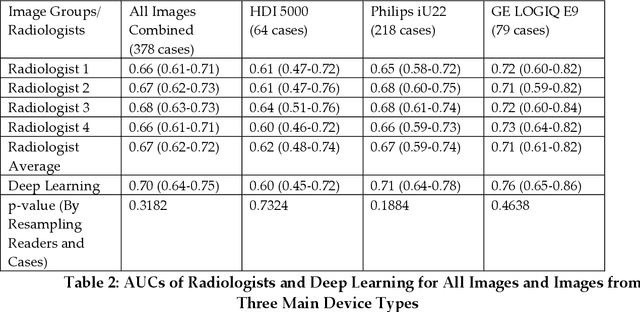
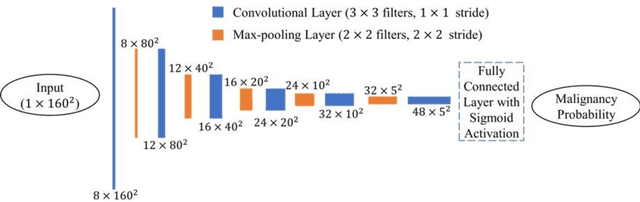
Abstract:Objectives: The purpose is to apply a previously validated deep learning algorithm to a new thyroid nodule ultrasound image dataset and compare its performances with radiologists. Methods: Prior study presented an algorithm which is able to detect thyroid nodules and then make malignancy classifications with two ultrasound images. A multi-task deep convolutional neural network was trained from 1278 nodules and originally tested with 99 separate nodules. The results were comparable with that of radiologists. The algorithm was further tested with 378 nodules imaged with ultrasound machines from different manufacturers and product types than the training cases. Four experienced radiologists were requested to evaluate the nodules for comparison with deep learning. Results: The Area Under Curve (AUC) of the deep learning algorithm and four radiologists were calculated with parametric, binormal estimation. For the deep learning algorithm, the AUC was 0.70 (95% CI: 0.64 - 0.75). The AUC of radiologists were 0.66 (95% CI: 0.61 - 0.71), 0.67 (95% CI:0.62 - 0.73), 0.68 (95% CI: 0.63 - 0.73), and 0.66 (95%CI: 0.61 - 0.71). Conclusion: In the new testing dataset, the deep learning algorithm achieved similar performances with all four radiologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge